Evaluating automatic cell number extraction from single cell papers
For the past ~7 years I have been tracking the scale of scRNA-seq experiments in a spreadsheet which we published as the ‘Single Cell Studies Database’ a few years ago (Svensson, da Veiga Beltrame, and Pachter 2020).
Initially, all fields were entered manually. Eventually, I realized that a lot of article information could be automatically populated from the article DOI. This is through a macro in Google Apps Script which queries crossref.org, receives an object containing the result data, pulls out the useful parts, formats them, and puts them in corresponding columns of the spreadsheet. This made it a lot easier to quickly add studies. This takes care of authors, journal, title, and publication date.
All other fields are still entered manually.
It is fascinating to see how the scale of studies are growing in size over the years. The genesis of tracking this trend was while writing a perspective article for Nature Protocols focused on technological development in the single cell genomics field, where we noted the exponential growth in scale for individual studies (Svensson, Vento-Tormo, and Teichmann 2018).
Over the years, I have learned some shortcuts for identifying the total cell numbers assayed for individual papers. Sometimes they are written in the abstract, often not. A quick search for the word ‘total’ in the paper can lead to sentences like ‘in total, we generated transcriptomes for 12,000 cells from liver and 20,000 cells from spleen’, then I can add them up to 22,000. If that doesn’t work, I tend to search for ‘cells’, which is obviously used everywhere in the paper, but the beginning of the ‘results’ section typically has numbers for collected cells. Sometimes the information is in figure legends. More often, it is just written as annotations in the figures themselves. For papers in more prestigious journals like Nature or Science, these kinds of details are more likely to be find in the supplemental material, which actually tend to a good thing, because those are usually available for free even if you don’t have access to the journal article itself. I also do some quick skimming of the paper, to see if I missed any way of getting these numbers that I didn’t think of. If I still can’t find it, I usually try to find the data accession ID, download the data, and try to see if I can easily load it and learn the numbers manually that way.
Recently, there has been an explosion of tools for data extraction from web sites using large language models. I wanted to try this strategy and see how far off they would be from my manually extracted cell numbers.
Evaluating Firecrawl for cell number extraction
The general pipeline is to have a scraper convert a query website into a local Markdown file, then pipe that file to an LLM together with a prompt to extract the information of interest. (It is healthy to keep in mind that a ‘paper’ is just a post on website by some guest authors, that happens to also have a more readable PDF version.)
There is a popular free tool called Crawl4AI, which has a handy CLI utility as well as a Python API. I played around with it for a bit, but while reading up on it I came across Firecrawl Extract.
In terms of functionality, Crawl4AI and Firecrawl Extract can do the same thing, with the key difference that you bring your own LLM API key or local LLM for a model of your choice to Crawl4AI, while this is built into extraction queries in Firecrawl Extract. For a quick test, I found Firecrawl Extract to be easier to get started with and getting results.
I sampled 97 scRNA-seq studies from the ‘Single Cell Studies Database’ where I had annotated the ‘Reported cells total’ column to use as an evaluation set.
Firecrawl Extract takes as input a URL, a prompt, and an output schema for structured outputs.
class ExtractSchema(BaseModel):
total_number_cells: int
failure_reason: str
url: str
prompt = '''
The page is a paper where the authors performed single-cell RNA-sequencing.
Extract the total number of cells the authors report to have collected.
Store the number in the total_number_cells field.
If you cannot extract the number of cells, give a brief reason in the failure_reason field.
Report the URL that was called to extract in the url field.
'''
Every DOI is a valid URL that resolves correctly when prepended with
https://doi.org/
. Even so, I had some hiccups where every ~14 papers or so my extraction loop exited with ValueError: ('Extract job failed. Error: All provided URLs are invalid. Please check your input and try again.', 500). Simply rerunning the query would result in a successful extraction job, so I’m not sure what was happening there. It could be due to rate limiting, but I put a pretty generous sleep.time(10) between each paper.
Results
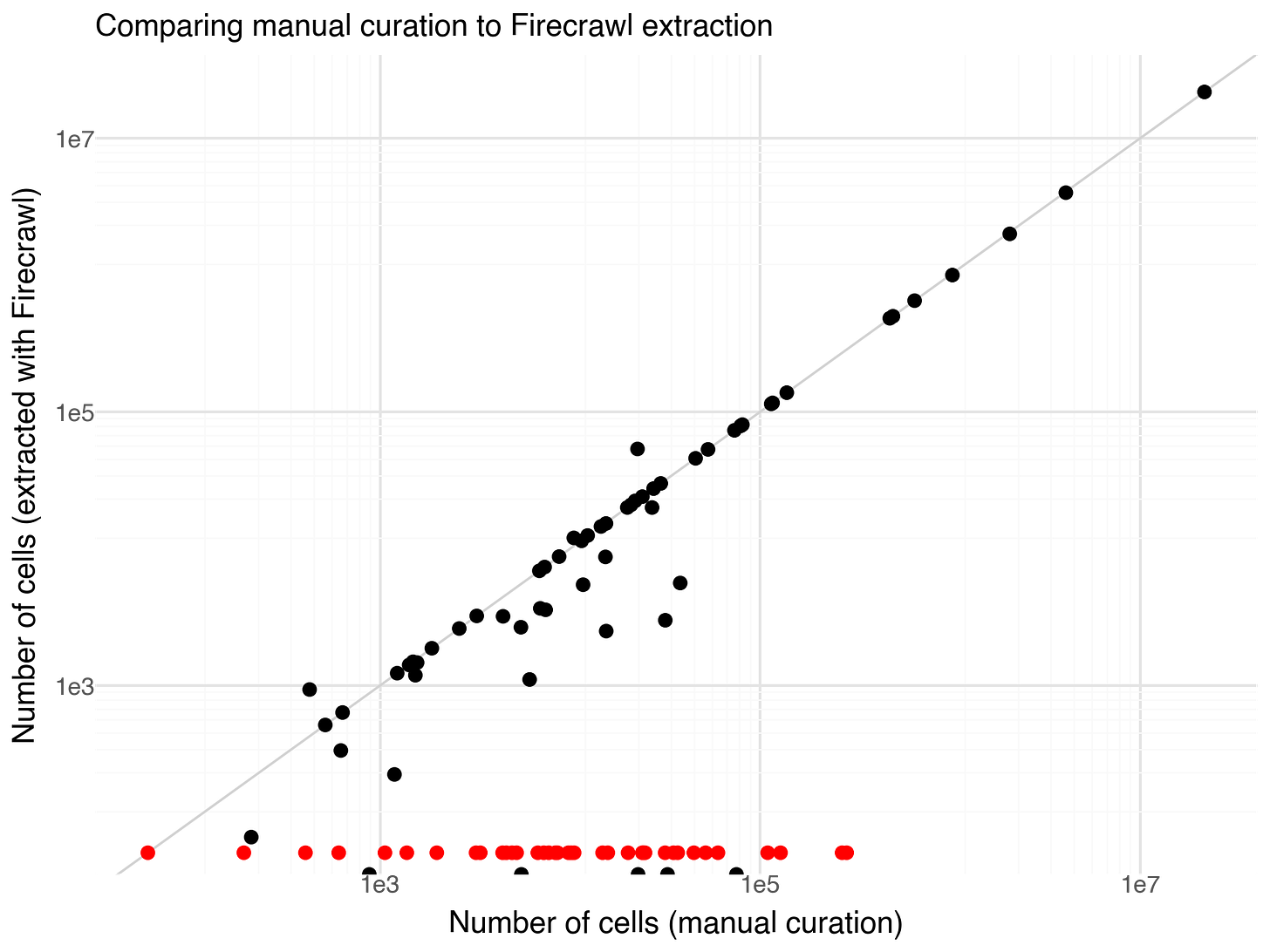
After extraction I could simply compare my manual annotations with the extractions from FireCrawl. In 38% of cases, Firecrawl Extract was not able to find a cell number and gave the reason that the number was not specified in the provided content. I suspect in most cases this is due to papers being behind paywalls. If I would rerun this experiment I would add further instructions to the prompt to report whether the URL has a paywall. Papers where Firecrawl Extract reported to not find cell numbers are included in the figure below as red dots.
The results had the exact same counts for 27% of the papers. Some of the 35% of papers with different cell counts can be explained by a few specific observations: a couple of the ‘Reported cells total’ values have very round numbers (e.g., 24,000) while Firecrawl Extract numbers or nearby but less round (e.g., 24,023). In these cases the manual curation is probably inaccurate. In some more cases the situation is reversed, for example, manual curation has 1,027,401 while Firecrawl reports 1,000,000. I think these are cases of Firecrawl picking up rounded numbers from the abstract (’In this study we profile a million cells …’ or so). There are a number of cases where Firecrawl reports 0 cells in the papers, but does not indicate that the information is missing. I’m not sure why this is happening.
Excluding cases where no cell numbers are reported, or numbers with 0 reported cells, the average fold change error in cell numbers was 1.4x.
The results are pretty binary. Either the extraction fails to find a cell number, or it is pretty likely to be near the actual number.
For now I think I will hold off on using this automation. I think the results are pretty useful, but the practical bottleneck is that if I was impromptu adding a paper to the spreadsheet, I would need to either use a CLI tool or go to the Firecrawl Extract dashboard and set up an extract job, and that is just too much friction.
I also like having confirmed manually curated numbers in the database for these sorts of evaluations. If I start using this automation I would want to add some kind of tag or additional column to indicate if the information was manually curated or automatically extracted.
Notebooks for this post are available on Github at https://github.com/vals/Blog/tree/master/250525-cell-number-extraction.
References
Svensson, Valentine, Eduardo da Veiga Beltrame, and Lior Pachter. 2020. “A Curated Database Reveals Trends in Single-Cell Transcriptomics.” Database: The Journal of Biological Databases and Curation 2020 (November). https://doi.org/10.1093/database/baaa073.
Svensson, Valentine, Roser Vento-Tormo, and Sarah A. Teichmann. 2018. “Exponential Scaling of Single-Cell RNA-Seq in the Past Decade.” Nature Protocols 13 (4): 599–604.