H5AD files in GEO
I’m working on a project where I want to use a lot of well-annotated scRNA-seq data. There are thousands of datasets available at GEO, but in the vast majority of cases these are basic outputs from CellRanger consisting of .mtx files with UMI counts and .tsv files with cell barcodes and gene names. That is, they are all missing experimental conditions and cell type labels.
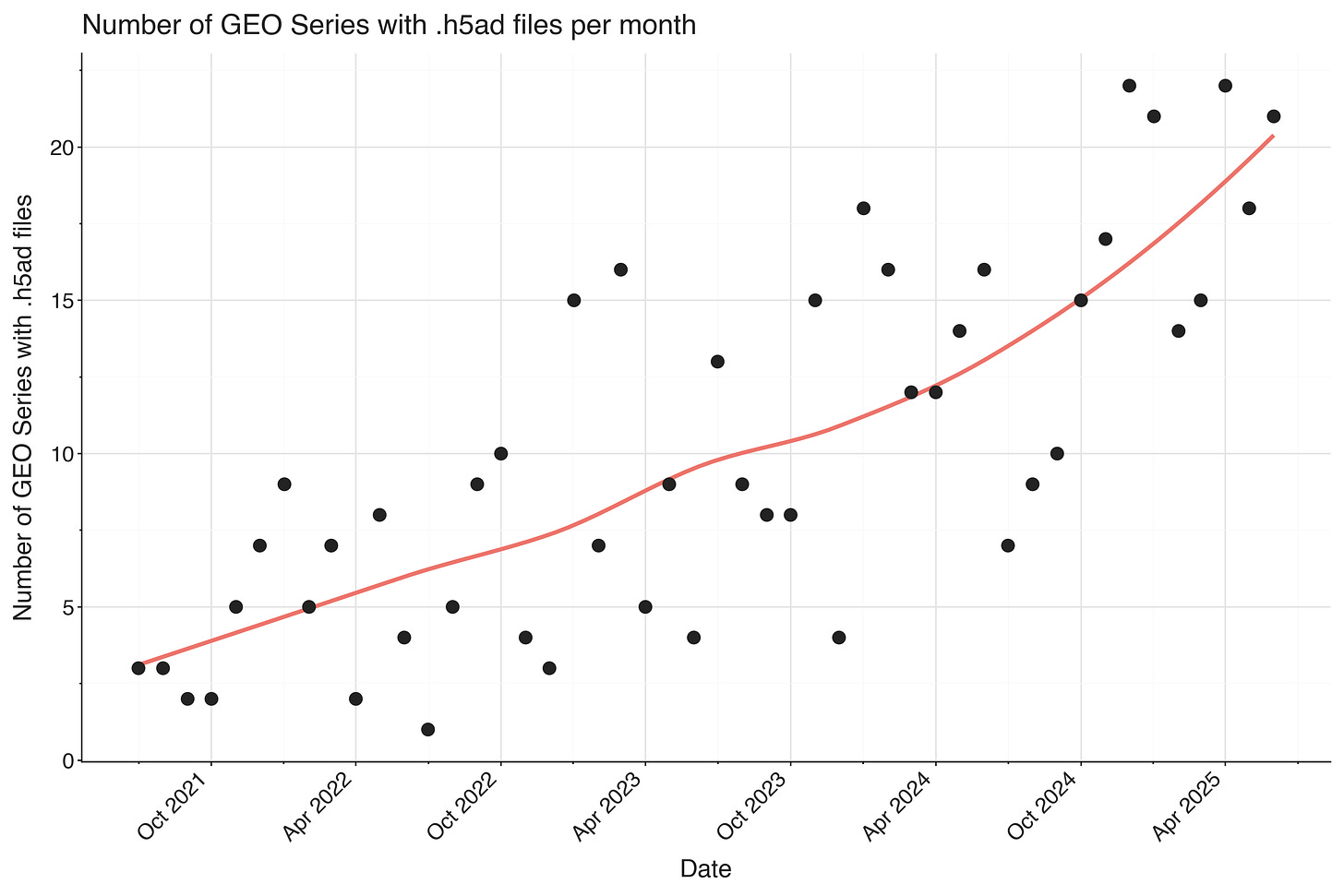
Some series on GEO have had anndata-based .h5ad files submitted. These are much more likely to have complete metadata and annotations included in them. For re-use, this is by far more valuable than the basic CellRanger output! In particular the cell type annotation process is very fraught and time consuming (you can usually infer experimental conditions from file names).
I was happily surprised that the number of GEO series with .h5ad files seem to be increasing over time!