Negative binomial regression and inference using a pre-trained transformer
I wanted to speed up negative binomial regression with a novel transformer model, but it actually turns out that a classical method of moments is a better solution. For this post I will focus on concepts, and instead refer to the resulting paper for more in depth details: https://arxiv.org/abs/2508.04111
Differences in counts
When analyzing scRNA-seq data, we are ultimately comparing molecule counts from different sources (genes, cell types, experimental conditions).
There are many other situations where your observations are counts.
As we learn to interpret data, we get good intuition about variabilities and magnitudes for measured values. These intuitions are typically based on more continuous values, and unfortunately counts behave differently from these.
Specifically, comparing magnitudes of observed counts between two groups of observations seems like it’s straightforward. Unfortunately, with a small number of samples (less than around ten) with observed counts of small magnitudes (also when typical counts are less than around ten) these comparisons are very challenging. Standard strategies for estimating means and variances from the data will fail, leading to inaccurate effect size estimation and p-values that can drastically change in magnitude from small sampling variation in the data.
The typical tool to solve these issues is negative binomial regression. This is used to estimate parameters needed to compare the groups of samples by directly modeling the properties of discrete counts. The negative binomial distribution has two parameters: mean and over-dispersion. Means are used for effect sizes. Over-dispersion indicates how much unidentified variation there is between samples on top of the variation from the counting process, which is needed to get p-values for the effect size.
Negative binomial regression is not magic though. Estimating the over-dispersion is very difficult with few observations and small counts. Such a small amount of data contains very limited information. Still, it is the best we have.
There is another issue however: the standard procedure to estimate the parameters of the negative binomial regression model is computationally expensive. For an individual comparison, computation is negligible, finishing in a millisecond. However, the ability to generate data through, for example, genome-wide CRISPR screens, and similar technologies, means that we need to do a huge number of comparisons. In the CRISPR screen example, if we knock down 20,000 genes and read out expression for 20,000 genes, we need to perform 400,000,000 comparisons. Even if a comparison takes a millisecond, this will still take over a hundred hours.
Iterative and non-iterative statistics
The reason that negative binomial regression is slow is because it typically uses an iterative method to estimate the parameters. It maximizes that likelihood by gradually improving the parameters estimates using analytical gradients.
These iterative parameter estimation methods have been around for a century, but were labor intensive before the invention of computers. A large amount of statistical methods research aimed to estimate parameters without iterative updates, referred to as non-iterative statistics. These methods use simple operations in one step to estimate statistics. For example, the mean in a normal distribution is non-iterative statistic; you sum the values and divide by the number of observations. You can estimate the mean with iterative maximum likelihood optimization, but it’s unnecessary, and might even produce worse results from numerical errors.
The standard iterative method to estimate the parameters in negative binomial distribution is called ‘iteratively reweighted least squares’.
Another way to estimate parameters is through the method of moments. Moments are values that describe properties of a probability distribution, such as mean, variance and skewness. For a given parameterized probability distribution, the observed moments can be expressed as functions of the parameters. In special cases, you can then solve for the parameters of interest with respect to the observed moments.
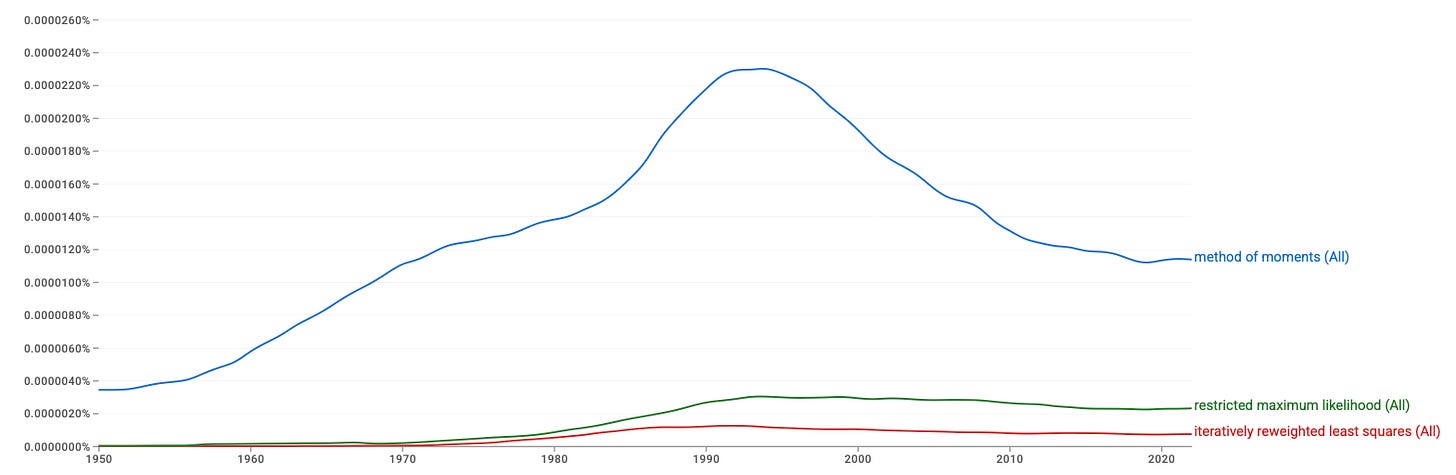
Up until the beginning of the 1990’s, there was increasing interest in the method of moments. In the early era of data collection, performing a statistical analysis was expensive. Puzzling out method of moments estimators for a given problem was a worthwhile effort, because the computational resources needed to do statistical inference were limited. As computational hardware became more affordable, the need for these custom solutions went away.
In addition to the work of finding method of moments estimators, maximum likelihood is optimally asymptotically efficient. It requires the the least possible amount of observations to bound the error of a parameter estimate within some interval. It is usually considered that method of moment estimation requires a larger number of observations for accurate results.
As our ability to collect data is outpacing performance improvements in computational hardware, we might want to revisit the concept of non-iterative statistics, to quickly get answers to the questions we have of the data.
Pair-set transformer for comparative statistics
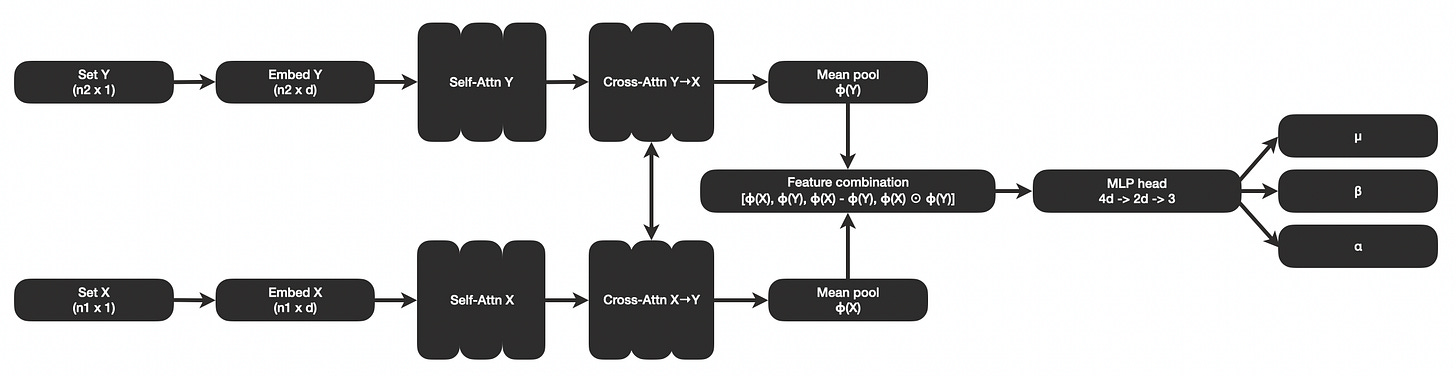
In a previous post I explored the idea of a pair-set transformer that can be pre-trained for the task of statistical estimation. The same architecture can be adapted to estimate the statistics needed for negative binomial regression and inference.
The general idea is to explore if modern machine learning frameworks such as transformers can be combined with the 1980’s concept of non-iterative statistics. Instead of deriving equations for particular special case problems, we can use a generic transformer-based neural network architecture combined with synthetic data generation to learn a network that predicts statistical parameters of interest.
While transformers are expensive to execute, relatively small ones can be more efficient than iterative parameter estimation. Training it will be expensive, but once it’s trained it can be re-used forever.
You simply show it the data, and it will predict what the statistical parameters of interest are.
The asymptotic optimality of maximum likelihood estimation means it would not be able to beat it in numeric performance, but may be a lot faster, and at least better than method of moments estimation.
I created a variation of the transformer architecture described in the previous post, designed for the task of estimating parameters needed to learn magnitude and significance in differences in counts. I trained the model and wrote up a paper with detailed methods and findings.
Method of moments actually performs really well
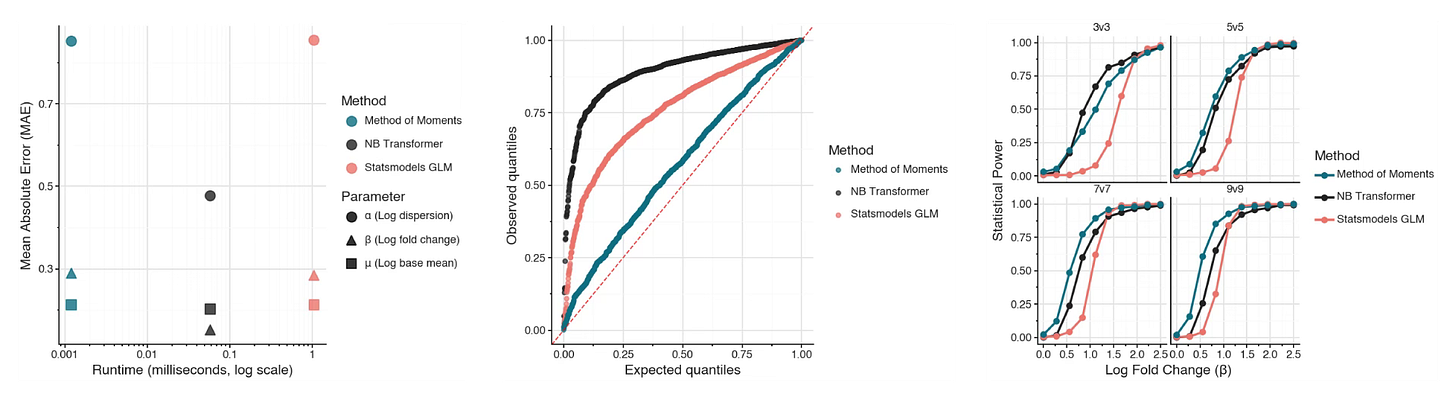
I compared the transformer-based method to the standard approach of estimating parameters through iterative maximum likelihood, as well as an analytical method of moments solution for the parameters. I did this because I figured the analytical solution would be extremely fast but have terrible performance.
It did not!
The method of moments solution had nearly identical estimation error as iterative maximum likelihood, much better P-value calibration than either the transformer method or the iterative method, and the most statistical power.
This was a big surprise for me.
For this analysis, I used practically realistic sample sizes: between two and ten replicates per condition. This is likely far below where the asymptotic optimality of iterative maximum likelihood starts being relevant.
While I found it interesting to work out the transformer architecture and training strategy, the more valuable part of this work was in the benchmarking. It identified that method of moments solution didn’t have the problems I expected it to have.
From around the web
Sequences and consequences - https://royalsocietypublishing.org/doi/10.1098/rstb.2009.0221
Optimizing murine sample sizes for RNA-seq studies revealed from large-scale comparative analysis - https://www.biorxiv.org/content/10.1101/2024.07.08.602525v1
A recap of virtual cell releases circa June 2025 - https://ekernf01.github.io/virtual-cell-june-2025
A bioinformatician, computer scientist, and geneticist lead bioinformatic tool development—which one is better? - https://academic.oup.com/bioinformaticsadvances/article/5/1/vbaf011/7989318
Which Kind of Science Reform - https://elevanth.org/blog/2025/07/09/which-kind-of-science-reform/
Using hierarchical modeling to get more stable rankings of gene expression - https://statmodeling.stat.columbia.edu/2025/07/30/using-hierarchical-modeling-to-get-more-stable-rankings-of-gene-expression/
(1) Fitting hierarchical models in genetics, (2) A Stan model that runs faster with 400,000 latent parameters, (3) Super-scalable penalized maximum likelihood inference for biome problems, (4) “In the end, I basically gave up working on biology bec... - https://statmodeling.stat.columbia.edu/2025/07/31/bio/
Overinterpreting underpowered multi-omics experiments - https://thecodon.substack.com/p/falling-in-the-trap-of-overinterpreting
The pharma industry from Paul Janssen to today: why drugs got harder to develop and what we can do about it - https://atelfo.github.io/2023/12/23/biopharma-from-janssen-to-today.html
The therapeutic potential of stem cells - https://royalsocietypublishing.org/doi/10.1098/rstb.2009.0149
Animals as chemical factories - https://worksinprogress.co/issue/animals-as-chemical-factories/
The immunology of asthma - https://www.nature.com/articles/s41590-025-02212-9
What's going on with gene therapies? (Part one) - https://nehalslearnings.substack.com/p/whats-going-on-with-gene-therapies
The Day Novartis Chose Discovery - https://www.alexkesin.com/p/the-day-novartis-chose-discovery
A Visual Guide to Gene Delivery - https://press.asimov.com/articles/gene-delivery
How pour-over coffee got good - https://worksinprogress.co/issue/how-pour-over-coffee-got-good/
THE LOTTERY OF FASCINATIONS - https://slatestarcodex.com/2013/06/30/the-lottery-of-fascinations/
The Truth Is Out There, Part 3: The Game of Belief - https://www.filfre.net/2024/10/the-truth-is-out-there-part-3-the-game-of-belief/
Alien: Earth (2025) - https://www.themoviedb.org/tv/157239-alien-earth
The immunology of asthma and chronic rhinosinusitis - https://www.nature.com/articles/s41577-025-01159-0
Hanseatic League - https://en.wikipedia.org/wiki/Hanseatic_League